Influence of Feature Selection and PCA on a Small Dataset
November 23, 2017
scikit-learn machine learning feature selection PCA cross-validationThis study covers the influence of feature selection and PCA on the Titanic Survivors dataset. Most of the preprocessing code such as data cleaning, encoding and transformation is adapted from the Scikit-Learn ML from Start to Finish work by Jeff Delaney.
Import Data
Load the csv train and test files into a pandas dataframe and print the first 5 rows to see a sample of the data. Print also a statistics description of each feature.
Show/Hide code
import pandas as pd
train_df = pd.read_csv("train.csv")
test_df = pd.read_csv("test.csv")
train_df.head()
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
Show/Hide code
train_df.describe()
| PassengerId | Survived | Pclass | Age | SibSp | Parch | Fare | |
|---|---|---|---|---|---|---|---|
| count | 891.000000 | 891.000000 | 891.000000 | 714.000000 | 891.000000 | 891.000000 | 891.000000 |
| mean | 446.000000 | 0.383838 | 2.308642 | 29.699118 | 0.523008 | 0.381594 | 32.204208 |
| std | 257.353842 | 0.486592 | 0.836071 | 14.526497 | 1.102743 | 0.806057 | 49.693429 |
| min | 1.000000 | 0.000000 | 1.000000 | 0.420000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 223.500000 | 0.000000 | 2.000000 | 20.125000 | 0.000000 | 0.000000 | 7.910400 |
| 50% | 446.000000 | 0.000000 | 3.000000 | 28.000000 | 0.000000 | 0.000000 | 14.454200 |
| 75% | 668.500000 | 1.000000 | 3.000000 | 38.000000 | 1.000000 | 0.000000 | 31.000000 |
| max | 891.000000 | 1.000000 | 3.000000 | 80.000000 | 8.000000 | 6.000000 | 512.329200 |
Visualise Data
To familiarise with the data and discover underlying patterns to exploit later in the machine learning models, we need to create some distribution, bar, and scatter plots. For a complete visualisation analysis check Scikit-Learn ML from Start to Finish work and my previous work here.
Engineer Features
- Aside from ‘Sex’, the ‘Age’ feature is second in importance. To avoid overfitting, group people into logical human age groups.
- Each ‘Cabin’ starts with a letter. Probably, this letter is more important than the number that follows, so slice it off.
- ‘Fare’ is another continuous value that should be simplified, placing the values into quartile bins accordingly.
- Extract information from the ‘Name’ feature. Rather than use the full name, extract the last name and prefix and then append them as their own features.
- Lastly, drop useless features (‘Ticket’, ‘Name’, and ‘Embarked’).
Show/Hide code
# Code adapted from https://www.kaggle.com/jeffd23/scikit-learn-ml-from-start-to-finish
def simplify_ages(df):
df.Age = df.Age.fillna(-0.5)
bins = (-1, 0, 5, 12, 18, 25, 35, 60, 120)
group_names = ['Unknown', 'Baby', 'Child', 'Teenager', 'Student', 'Young Adult', 'Adult', 'Senior']
categories = pd.cut(df.Age, bins, labels=group_names)
df.Age = categories
return df
def simplify_cabins(df):
df.Cabin = df.Cabin.fillna('N')
df.Cabin = df.Cabin.apply(lambda x: x[0])
return df
def simplify_fares(df):
df.Fare = df.Fare.fillna(-0.5)
bins = (-1, 0, 8, 15, 31, 1000)
group_names = ['Unknown', '1_quartile', '2_quartile', '3_quartile', '4_quartile']
categories = pd.cut(df.Fare, bins, labels=group_names)
df.Fare = categories
return df
def format_name(df):
df['Lname'] = df.Name.apply(lambda x: x.split(' ')[0])
df['NamePrefix'] = df.Name.apply(lambda x: x.split(' ')[1])
return df
def drop_features(df):
return df.drop(['Ticket', 'Name', 'Embarked'], axis=1)
def transform_features(df):
df = simplify_ages(df)
df = simplify_cabins(df)
df = simplify_fares(df)
df = format_name(df)
df = drop_features(df)
return df
train_df = transform_features(train_df)
test_df = transform_features(test_df)
train_df.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Fare | Cabin | Lname | NamePrefix | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | male | Student | 1 | 0 | 1_quartile | N | Braund, | Mr. |
| 1 | 2 | 1 | 1 | female | Adult | 1 | 0 | 4_quartile | C | Cumings, | Mrs. |
| 2 | 3 | 1 | 3 | female | Young Adult | 0 | 0 | 1_quartile | N | Heikkinen, | Miss. |
| 3 | 4 | 1 | 1 | female | Young Adult | 1 | 0 | 4_quartile | C | Futrelle, | Mrs. |
| 4 | 5 | 0 | 3 | male | Young Adult | 0 | 0 | 2_quartile | N | Allen, | Mr. |
Encode Data
Normalize and transform categorical non-numerical features to numerical with the LabelEncoder tool from scikit-learn, making out data more flexible for various algorithms.
Show/Hide code
from sklearn import preprocessing
def encode_features(df_train, df_test):
features = ['Fare', 'Cabin', 'Age', 'Sex', 'Lname', 'NamePrefix']
df_combined = pd.concat([df_train[features], df_test[features]])
for feature in features:
le = preprocessing.LabelEncoder()
le = le.fit(df_combined[feature])
df_train[feature] = le.transform(df_train[feature])
df_test[feature] = le.transform(df_test[feature])
return df_train, df_test
train_df, test_df = encode_features(train_df, test_df)
train_df.head()
| PassengerId | Survived | Pclass | Sex | Age | SibSp | Parch | Fare | Cabin | Lname | NamePrefix | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | 1 | 4 | 1 | 0 | 0 | 7 | 100 | 19 |
| 1 | 2 | 1 | 1 | 0 | 0 | 1 | 0 | 3 | 2 | 182 | 20 |
| 2 | 3 | 1 | 3 | 0 | 7 | 0 | 0 | 0 | 7 | 329 | 16 |
| 3 | 4 | 1 | 1 | 0 | 7 | 1 | 0 | 3 | 2 | 267 | 20 |
| 4 | 5 | 0 | 3 | 1 | 7 | 0 | 0 | 1 | 7 | 15 | 19 |
Machine Learning
Split Data to Train/Test Sets
Create train/test sets using the train_test_split function. The test_size=0.2 indicates the percentage of the data that should be held over for testing.
Show/Hide code
from sklearn.model_selection import train_test_split
# Define the independent variables as features.
Xs = train_df.drop(['PassengerId', 'Survived'], axis=1)
# Define the target (dependent) variable as labels.
Ys = train_df['Survived']
# Create a train/test split using 30% test size.
X_train, X_test, y_train, y_test = train_test_split(Xs,
Ys,
test_size=0.2,
random_state=23)
# Check the split printing the shape of each set.
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
(712, 9) (712,)
(179, 9) (179,)
Evaluate Algorithms
Test and evaluate 5 algorithms: 1. Naive Bayes 2. Support Vector Machines 3. K Nearest Neighbors 4. Random Forest 5. AdaBoost
Validate
Measure the effectiveness of the algorithm applying KFold. Split the data into 50 buckets, then run the algorithm using a different bucket as the test set for each iteration. Turn shuffle=True to shuffle the data points’ order before splitting into folds.
Show/Hide code
import numpy as np
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.model_selection import KFold
from time import time
# Create the classifiers.
nb = GaussianNB()
svc = SVC()
dtc = DecisionTreeClassifier()
knc = KNeighborsClassifier()
rfc = RandomForestClassifier()
abc = AdaBoostClassifier()
# Create a dictionary of classifiers to choose from.
classifiers = {"GaussianNB": nb, "SVM": svc, "Decision Trees": dtc,
"KNN": knc, "Random Forest": rfc, "AdaBoost": abc}
# Create a function that runs and evaluates the classifiers.
def test_clfs(clf):
# Create the KFold cross validation iterator.
kf = KFold(n_splits=50, shuffle=True, random_state=23)
outcomes = []
fold = 0
for train_index, test_index in kf.split(Xs):
t0 = time()
fold += 1
X_train, X_test = Xs.values[train_index], Xs.values[test_index]
y_train, y_test = Ys.values[train_index], Ys.values[test_index]
# Fit the classifier to the data.
clf.fit(X_train, y_train)
# Create a set of predictions.
predictions = clf.predict(X_test)
# Evaluate predictions with accuracy score.
accuracy = clf.score(X_test, y_test)
outcomes.append(accuracy)
mean_outcome = np.mean(outcomes)
# Print the results.
print("\nMean Accuracy: {0}".format(mean_outcome))
print("\nTime passed: ", round(time() - t0, 3), "s\n")
for name, clf in classifiers.items():
print("#"*55)
print(name)
test_clfs(clf)
#######################################################
GaussianNB
Mean Accuracy: 0.7601960784313725
Time passed: 0.002 s
#######################################################
SVM
Mean Accuracy: 0.6271895424836601
Time passed: 0.03 s
#######################################################
Decision Trees
Mean Accuracy: 0.7644444444444445
Time passed: 0.002 s
#######################################################
KNN
Mean Accuracy: 0.5996078431372549
Time passed: 0.002 s
#######################################################
Random Forest
Mean Accuracy: 0.8041176470588235
Time passed: 0.02 s
#######################################################
AdaBoost
Mean Accuracy: 0.8263398692810456
Time passed: 0.088 s
- Select Adaboost as the best algorithm, since it gives the best accuracy scores.
Influence of Feature Selection & PCA
Investigate if Feature Selection and PCA can improve the performance of Random Forest.
Feature Selection
Select the best features with SelectKBest, which removes all but the k highest scoring features.
PCA
Reduce the dimensionnality of the data using the Principal Component Analysis (PCA).
Dimensionality Reduction
Use GridSearchCV and Pipeline to optimize over different classes of estimators. Compare unsupervised PCA dimensionality reduction to univariate feature selection SelectKBest during the grid search.
Show/Hide code
from sklearn.feature_selection import SelectKBest
from sklearn.decomposition import PCA
from sklearn.model_selection import GridSearchCV, KFold
from sklearn.pipeline import Pipeline
from sklearn.metrics import make_scorer, accuracy_score, classification_report, confusion_matrix
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# Create the classifier.
clf = AdaBoostClassifier()
# Create the pipeline.
pipeline = Pipeline([('reduce_dim', PCA()),
('clf', clf)])
# Create the parameters.
n_feature_options = [1, 2, 3, 4, 5, 6, 7, 8, 9]
n_estimators = [50]
parameters = [{'reduce_dim': [PCA(iterated_power=7)],
'reduce_dim__n_components': n_feature_options,
'clf__n_estimators': n_estimators},
{'reduce_dim': [SelectKBest()],
'reduce_dim__k': n_feature_options,
'clf__n_estimators': n_estimators}]
reducer_labels = ['PCA', 'KBest()']
# Create a function to get the best estimator and print the reports.
def compare_estimators():
t0 = time()
# Create the KFold cross-validator.
kf = KFold(n_splits=50, shuffle=True, random_state=23)
# Create accuracy score to compare each combination.
scoring = {'Accuracy': make_scorer(accuracy_score)}
# Create the grid search.
grid = GridSearchCV(estimator=pipeline,
param_grid=parameters,
scoring=scoring,
cv=kf, refit='Accuracy')
# Fit grid search combinations.
grid.fit(X_train, y_train)
# Make predictions.
predictions = grid.predict(X_test)
# Evaluate using sklearn.classification_report().
report = classification_report(y_test, predictions)
# Get the best parameters and scores.
best_parameters = grid.best_params_
best_score = grid.best_score_
mean_scores = np.array(grid.cv_results_['mean_test_Accuracy'])
# scores are in the order of param_grid iteration, which is alphabetical
mean_scores = mean_scores.reshape(len(n_estimators), -1, len(n_feature_options))
# select score for best C
mean_scores = mean_scores.max(axis=0)
bar_offsets = (np.arange(len(n_feature_options)) *
(len(reducer_labels) + 1) + .5)
plt.figure(figsize=(10, 5))
for i, (label, reducer_scores) in enumerate(zip(reducer_labels, mean_scores)):
plt.bar(bar_offsets + i, reducer_scores, label=label)
plt.title("Comparing feature reduction techniques")
plt.xlabel('Reduced number of features')
plt.xticks(bar_offsets + len(reducer_labels) / 2, n_feature_options)
plt.ylabel('Accuracy')
plt.ylim((0, 1))
plt.legend(loc='upper left')
# Print the results.
print("\nAccuracy score: ", accuracy_score(y_test, predictions))
print("\nReport:\n")
print(report)
print("\nBest Mean Accuracy score: ", best_score)
print("\nBest parameters:\n")
print(best_parameters)
print(confusion_matrix(y_test, predictions))
print("Time passed: ", round(time() - t0, 3), "s")
return grid.best_estimator_
compare_estimators()
Accuracy score: 0.821229050279
Report:
precision recall f1-score support
0 0.85 0.87 0.86 115
1 0.76 0.73 0.75 64
avg / total 0.82 0.82 0.82 179
Best Mean Accuracy score: 0.817415730337
Best parameters:
{'clf__n_estimators': 50, 'reduce_dim': SelectKBest(k=9, score_func=<function f_classif at 0x116dfee18>), 'reduce_dim__k': 9}
[[100 15]
[ 17 47]]
Time passed: 101.808 s
Pipeline(memory=None,
steps=[('reduce_dim', SelectKBest(k=9, score_func=<function f_classif at 0x116dfee18>)), ('clf', AdaBoostClassifier(algorithm='SAMME.R', base_estimator=None,
learning_rate=1.0, n_estimators=50, random_state=None))])
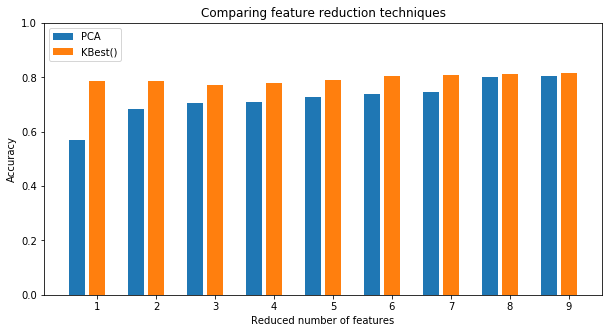
- It seems that
SelectKBestperforms better thanPCAfor all the cases, but the accuracy score has not improved significantly after dimensionality reduction. Actually, it was found that the best model keeps all the features (k=9).
Tune
To get an improved performance, optimise the hyperparameters that impact the model using GridSearchCV. The effectiveness of the algorithm is validated with StratifiedShuffleSplit and the evaluation with multiple metrics such as accuracy, precision, and recall.
Show/Hide code
from sklearn.model_selection import StratifiedShuffleSplit
# Create the classifier.
clf = AdaBoostClassifier()
# Create the parameters.
parameters = {'n_estimators': [10, 25, 50, 75],
'algorithm': ['SAMME', 'SAMME.R'],
'random_state': [3]}
# Find the best estimator and print the reports.
t0 = time()
# Create the Stratified ShuffleSplit cross-validator.
sss = StratifiedShuffleSplit(n_splits=50, test_size=0.2, random_state=3)
# Create multiple evaluation metrics to compare each combination.
scoring = {'AUC': 'roc_auc',
'Accuracy': make_scorer(accuracy_score),
'Precision': 'precision',
'Recall': 'recall',
'f1': 'f1'}
# Create the grid search.
grid = GridSearchCV(estimator=clf,
param_grid=parameters,
scoring=scoring,
cv=sss, refit='Accuracy')
# Fit grid search combinations.
grid.fit(X_train, y_train)
# Make predictions.
predictions = grid.predict(X_test)
# Evaluate using sklearn.classification_report().
report = classification_report(y_test, predictions)
# Get the best parameters and scores.
best_parameters = grid.best_params_
best_score = grid.best_score_
# Print the results.
print("\nAccuracy score: ", accuracy_score(y_test, predictions))
print("\nReport:\n")
print(report)
print("\nBest Accuracy score: ", best_score)
print("\nBest parameters:\n")
print(best_parameters)
print(confusion_matrix(y_test, predictions))
print("Time passed: ", round(time() - t0, 3), "s")
best_clf = grid.best_estimator_
Accuracy score: 0.821229050279
Report:
precision recall f1-score support
0 0.85 0.87 0.86 115
1 0.76 0.73 0.75 64
avg / total 0.82 0.82 0.82 179
Best Accuracy score: 0.819300699301
Best parameters:
{'algorithm': 'SAMME.R', 'n_estimators': 25, 'random_state': 3}
[[100 15]
[ 17 47]]
Time passed: 42.804 s
Predict the Actual Test Data
Finally, make the predictions and export them to a csv file.
Show/Hide code
passenger_ids = test_df['PassengerId']
predictions = best_clf.predict(test_df.drop('PassengerId', axis=1))
output = pd.DataFrame({ 'PassengerId' : passenger_ids, 'Survived': predictions })
output.to_csv('titanic_predictions.csv', index = False)
output.head()
| PassengerId | Survived | |
|---|---|---|
| 0 | 892 | 0 |
| 1 | 893 | 0 |
| 2 | 894 | 0 |
| 3 | 895 | 0 |
| 4 | 896 | 1 |